Source: The Conversation (Au and NZ) – By David Welch, Senior lecturer, University of Auckland
Genetic surveillance — a technology that uses the genetic fingerprint of a virus to track its spread — is part of the public health response to New Zealand’s new COVID-19 community outbreak and could help pinpoint its source.
There are now 17 cases of community transmission, all in Auckland, and health officials are treating the group as a single cluster, with an expectation that case numbers will grow.
Ideally, we should be sequencing all positive test swabs, regardless of whether they are found at the border or in the community. The community cases could then be compared to all other cases to find a close match. This would suggest a likely chain of transmission, help with contact tracing and reveal the sequence of the outbreak.
But not all samples are currently sequenced. In total, New Zealand had 1225 confirmed cases of COVID-19, and about 700 of the positive samples have been sequenced.
I argue the Ministry of Health should now make genetic sequencing mandatory. Here’s why.Tracking epidemics using genomes
Genetic surveillance of infectious diseases is a maturing technology that has played a major role in the effort to control the Ebola and Zika epidemics, and now the COVID-19 pandemic.
We can now obtain a complete viral genome quickly and cheaply to identify mutations that provide clues about transmission chains.
Cases that are part of the same transmission chain will have genomes that look very similar: they share the same mutations compared to a reference genome. Cases from different transmission chains have genomes with differing patterns of mutations.

SARS-CoV-2, the virus that causes COVID-19, does not have a particularly high mutation rate. It acquires mutations at about half the rate of seasonal influenza, but it mutates fast enough to leave a signal of where it has come from.
Read more: ‘Genomic fingerprinting’ helps us trace coronavirus outbreaks. What is it and how does it work?
This brings us to how this is helping in our efforts to control the current community outbreak.
There are four main theories about where the new cases could have come from:
The genomes of the new cases could identify the first scenario of a quarantine leak if we found a match between viral genomes from people in quarantine and in the new cluster. This relies on comprehensive sequencing of all cases in quarantine, but currently, there are still gaps.
Similarly, in the unlikely scenario of a transmission chain that has persisted since New Zealand’s first wave, we’d expect a match with one of the cases sequenced during the first outbreak, although the genomes would have diverged somewhat over that period of time.
The scenarios of transmission through goods or an undetected border case are more difficult to decipher using genomic methods. We would be looking to match the viral genome from the new local cases to one of more than 80,000 publicly available genomes that have been sampled worldwide. This would point to a country of origin but not necessarily distinguish between the scenarios.
Early results from sequencing of the first four cases from the new Auckland cluster suggest no link to a known (sequenced) New Zealand case, and the UK as the closest match. For now, this leaves all possibilities still open.
Ongoing surveillance
With widespread testing now underway, new cases will be identified in the community over the coming weeks. It is important that they are rapidly sequenced to determine whether they belong to the same transmission chain.
Genomic analysis will tell us whether we are dealing with a single or multiple clusters. Even the best contact tracing cannot be sure of the origin of an infection, and supplementing it with genomic data is crucial.
But genomic analysis is not limited to establishing transmission chains. It can also tell us about the overall size of an outbreak, which is directly related to the genomic diversity of the virus. We can also date events to establish when transmission started within a cluster, provided there is sufficient diversity in the cluster.
The genomes we have so far in New Zealand show a huge diversity of cases, with many introductions from around the globe. Indeed, the diversity of early samples largely reflects the diversity of the virus globally, with most cases that led to further transmission coming from North America and Australia in line with travel patterns to New Zealand.
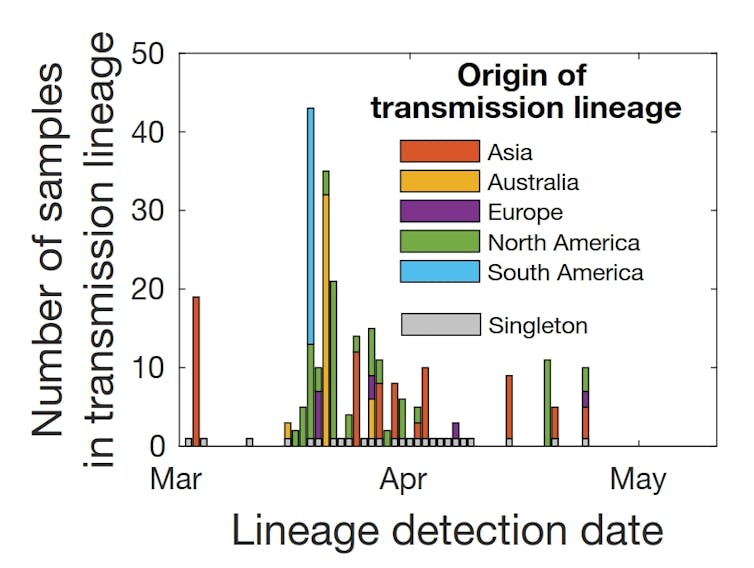
Most introductions did not result in further community transmission. This shows how effective New Zealand’s first lockdown was, when transmission rates declined dramatically soon after level 3 and 4 measures were put in place.
Genomes were also used in real time during the first outbreak to help attribute cases to clusters. Retrospectively, this has shown that contact tracing was effective, with relatively few cases being wrongly attributed.
But genome analysis is neither foolproof nor a panacea. Sometimes positive samples are found that cannot be sequenced because they contain only a small amount of viral material. The rather slow rate of mutation of Sars-COV-2 means many cases are essentially carrying identical copies of the virus, even across different countries.
This greatly reduces our ability to attribute an infection to a particular outbreak. There are also real computational bottlenecks – data is generated faster than we can sensibly analyse it.
Despite these limitations, genomic surveillance gives us near real-time insights into the spread of COVID-19 that were not possible in any previous pandemic. That’s why I argue it’s time for the Ministry of Health to now make immediate genetic sequencing mandatory for all positive test swabs in New Zealand, not just some.
– ref. Why New Zealand needs to focus on genome sequencing to trace the source of its new COVID-19 outbreak – https://theconversation.com/why-new-zealand-needs-to-focus-on-genome-sequencing-to-trace-the-source-of-its-new-covid-19-outbreak-144402